class: center, middle, inverse, title-slide # Restructuring Data --- layout: true <div class="dk-footer"> <span> <a href="https://rfortherestofus.com/" target="_blank">R for the Rest of Us </a> </span> </div> --- class: center, middle, dk-section-title background-image:url("images/casey-horner-KtlTvi8leYQ-unsplash.jpg") background-size: cover # Toolbox Refresher ??? In previous lessons, we defined data cleaning and learned how to organize our data. Now we can get started with using R, and in particular, on first getting our data into sensible, usable structures. Let’s begin by going over the tools and packages that we’ll be using for this part and throughout this course. --- ### Tools for preparing, transforming, and restructuring data ??? When working in R for data cleaning, we’ll be calling on functions from two popular packages: dplyr and tidyr. Both are written and maintained by Rstudio staff with extensive community contributions. They are part of larger set of packages referred to as the tidyverse. -- .fl.w-40[ .tc[ 📦 .large.b.rrured[`dplyr`] 📦 .large.b.rrured[`tidyr`] ]] .fl.w-60[ .large[Part of the .b.rrured[`tidyverse`]] Flexible, but developed around built-in features that support a particular way of doing things ] ??? These tools are meant to be flexible, but they are built around built-in features that support a particular way of doing specific tasks. -- - Hard to use incorrectly by accident - Less code to achieve a goal - Shared grammar and input/output data structures ??? Functions in different tidyverse packages follow tidy data principles so they work well together. And, by having specialized purposes, each one does only one thing but they do it well. Having established that this course is tidyverse-oriented. Let’s go over another tool that we’ll be using to make our code easy to read. --- ## Pipes To perform multiple operations in sequence: ??? When we work with functions to perform many sequential operations, we have three options. -- - Nested code ??? we can use nested code, in which function calls are nested inside more function calls so that the results get evaluated from the inside out. -- - Intermediate objects ??? We can also assign objects with intermediate results, passed on to the next function in our sequence. -- - Pipes ??? Or, as we’ll be doing in this course, we can use pipes. Let me show you how this looks like. --- ## Nested code ```r grades <- c(7,8,8,10,6,9,5,9,8) round(mean(grades), digits = 2) ``` ``` ## [1] 7.78 ``` ??? Nested code looks like this. Here, we have a vector ‘grades’ with some numeric values. Our sequence of operations is to first get the mean grade, and then round this result. In nested code, the mean function is nested inside the round function. -- ## Intermediate objects ```r mn_grades <- mean(grades) rd_mn_grades <- round(mn_grades, digits = 2) rd_mn_grades ``` ``` ## [1] 7.78 ``` ??? Intermediate objects work in a similar way. First we assign an object with the mean of the grades, then we pass this object to the rounding function Notice how both approaches give us the same result. --- ## Pipes For structuring sequential operations left-to-right: .fl.w-third[ .bt.bb[ Left Hand Side (.avenir[LHS]) <br> ]] .fl.w-third[ .bt.bb[ `pipe operator` .b.orange[`%>%`] from 📦 .b.rrured[`magrittr`] ]] .fl.w-third[ .bt.bb[ Right Hand Side (.avenir[RHS]) <br> ]] ??? Alternatively, we can use pipes to avoid nested code or having so assign results to intermediate objects that we don’t really need. Pipes let us structure sequential operations from left to right. When we work with pipes, there is always something on the left hand side of a pipe operator, and something else on the right hand side. The pipe operator appears in orange here. Its originally from the magrittr package --- ## Pipes - Take an **object** on the .avenir[Left Hand Side] of the pipe and insert it into a **function** as an argument on the .avenir[Right Hand Side] ??? More specifically, pipes take and object on the left hand side of the pipe operator and insert it into a function on the right hand side. -- - By default, .avenir[LHS] is placed as the **first** argument in the call ??? By default, the object on the left hand side becomes the first argument in the function call. -- - Packages in the `tidyverse` load .b.orange[`%>%`] automatically ??? by the way, the pipe operator gets loaded by the tidyverse packages that we’ll be using. --- ## Pipes ??? Let me summarize the main advantages of using pipes to write code. -- - Minimize the need for intermediate objects and nested code ??? First, by chaining our operations with pipes we pass the results onto the next function, so we don’t need nested code or to assign intermediate results to objects we won’t use again. -- - Make code readable ??? This makes our code more readable, and -- - Easy to add or remove steps at any point in the sequence of operations ??? its also easier to add or remove steps in our sequence of operations. -- > Insert with `ctrl + shift + M` ??? A useful shortcut to remember is that in Rstudio we can insert pipes using control shift M, or command shift m on a mac. -- ```r library(magrittr) grades %>% mean() %>% round(digits=2) ``` ``` ## [1] 7.78 ``` ??? The example from earlier looks like this is we use pipes. First we load the magrittr package, take the grades object, pass it to the mean function, and then pass this result to the round function. Note how we don’t have to write the first argument in functions that are on the right hand side of a pipe. We can now learn about the functions from dplyr that we’ll use extensively throughout this course. --- ## Useful 📦 .b.rrured[`dplyr`] functions ??? As I mentioned earlier, the dplyr package provides us with separate functions for doing the most common data manipulation tasks. -- - Individual functions for the most common operations ??? dplyr provides separate functions for the more common operations -- - Each function does one only thing, but does it well ??? Each function does one only thing, but does it well -- - Intuitive, user-friendly functions for common data manipulation tasks: ??? When we’re cleaning data, we usually do one of these if not all three of the following: -- - Subset columns ??? subset columns -- - Subset rows ??? Subset rows -- - Create or modify columns ??? or create or modify columns --- .panelset[ .panel[.panel-name[beers] <style>html { font-family: -apple-system, BlinkMacSystemFont, 'Segoe UI', Roboto, Oxygen, Ubuntu, Cantarell, 'Helvetica Neue', 'Fira Sans', 'Droid Sans', Arial, sans-serif; } #vcbkhkbzey .gt_table { display: table; border-collapse: collapse; margin-left: auto; margin-right: auto; color: #333333; font-size: 16px; font-weight: normal; font-style: normal; background-color: #FFFFFF; width: auto; border-top-style: solid; border-top-width: 2px; border-top-color: #A8A8A8; border-right-style: none; border-right-width: 2px; border-right-color: #D3D3D3; border-bottom-style: solid; border-bottom-width: 2px; border-bottom-color: #A8A8A8; border-left-style: none; border-left-width: 2px; border-left-color: #D3D3D3; } #vcbkhkbzey .gt_heading { background-color: #FFFFFF; text-align: center; border-bottom-color: #FFFFFF; border-left-style: none; border-left-width: 1px; border-left-color: #D3D3D3; border-right-style: none; border-right-width: 1px; border-right-color: #D3D3D3; } #vcbkhkbzey .gt_title { color: #333333; font-size: 125%; font-weight: initial; padding-top: 4px; padding-bottom: 4px; border-bottom-color: #FFFFFF; border-bottom-width: 0; } #vcbkhkbzey .gt_subtitle { color: #333333; font-size: 85%; font-weight: initial; padding-top: 0; padding-bottom: 4px; border-top-color: #FFFFFF; border-top-width: 0; } #vcbkhkbzey .gt_bottom_border { border-bottom-style: solid; border-bottom-width: 2px; border-bottom-color: #D3D3D3; } #vcbkhkbzey .gt_col_headings { border-top-style: solid; border-top-width: 2px; border-top-color: #D3D3D3; border-bottom-style: solid; border-bottom-width: 2px; border-bottom-color: #D3D3D3; border-left-style: none; border-left-width: 1px; border-left-color: #D3D3D3; border-right-style: none; border-right-width: 1px; border-right-color: #D3D3D3; } #vcbkhkbzey .gt_col_heading { color: #333333; background-color: #FFFFFF; font-size: 100%; font-weight: normal; text-transform: inherit; border-left-style: none; border-left-width: 1px; border-left-color: #D3D3D3; border-right-style: none; border-right-width: 1px; border-right-color: #D3D3D3; vertical-align: bottom; padding-top: 5px; padding-bottom: 6px; padding-left: 5px; padding-right: 5px; overflow-x: hidden; } #vcbkhkbzey .gt_column_spanner_outer { color: #333333; background-color: #FFFFFF; font-size: 100%; font-weight: normal; text-transform: inherit; padding-top: 0; padding-bottom: 0; padding-left: 4px; padding-right: 4px; } #vcbkhkbzey .gt_column_spanner_outer:first-child { padding-left: 0; } #vcbkhkbzey .gt_column_spanner_outer:last-child { padding-right: 0; } #vcbkhkbzey .gt_column_spanner { border-bottom-style: solid; border-bottom-width: 2px; border-bottom-color: #D3D3D3; vertical-align: bottom; padding-top: 5px; padding-bottom: 6px; overflow-x: hidden; display: inline-block; width: 100%; } #vcbkhkbzey .gt_group_heading { padding: 8px; color: #333333; background-color: #FFFFFF; font-size: 100%; font-weight: initial; text-transform: inherit; border-top-style: solid; border-top-width: 2px; border-top-color: #D3D3D3; border-bottom-style: solid; border-bottom-width: 2px; border-bottom-color: #D3D3D3; border-left-style: none; border-left-width: 1px; border-left-color: #D3D3D3; border-right-style: none; border-right-width: 1px; border-right-color: #D3D3D3; vertical-align: middle; } #vcbkhkbzey .gt_empty_group_heading { padding: 0.5px; color: #333333; background-color: #FFFFFF; font-size: 100%; font-weight: initial; border-top-style: solid; border-top-width: 2px; border-top-color: #D3D3D3; border-bottom-style: solid; border-bottom-width: 2px; border-bottom-color: #D3D3D3; vertical-align: middle; } #vcbkhkbzey .gt_from_md > :first-child { margin-top: 0; } #vcbkhkbzey .gt_from_md > :last-child { margin-bottom: 0; } #vcbkhkbzey .gt_row { padding-top: 8px; padding-bottom: 8px; padding-left: 5px; padding-right: 5px; margin: 10px; border-top-style: solid; border-top-width: 1px; border-top-color: #D3D3D3; border-left-style: none; border-left-width: 1px; border-left-color: #D3D3D3; border-right-style: none; border-right-width: 1px; border-right-color: #D3D3D3; vertical-align: middle; overflow-x: hidden; } #vcbkhkbzey .gt_stub { color: #333333; background-color: #FFFFFF; font-size: 100%; font-weight: initial; text-transform: inherit; border-right-style: solid; border-right-width: 2px; border-right-color: #D3D3D3; padding-left: 12px; } #vcbkhkbzey .gt_summary_row { color: #333333; background-color: #FFFFFF; text-transform: inherit; padding-top: 8px; padding-bottom: 8px; padding-left: 5px; padding-right: 5px; } #vcbkhkbzey .gt_first_summary_row { padding-top: 8px; padding-bottom: 8px; padding-left: 5px; padding-right: 5px; border-top-style: solid; border-top-width: 2px; border-top-color: #D3D3D3; } #vcbkhkbzey .gt_grand_summary_row { color: #333333; background-color: #FFFFFF; text-transform: inherit; padding-top: 8px; padding-bottom: 8px; padding-left: 5px; padding-right: 5px; } #vcbkhkbzey .gt_first_grand_summary_row { padding-top: 8px; padding-bottom: 8px; padding-left: 5px; padding-right: 5px; border-top-style: double; border-top-width: 6px; border-top-color: #D3D3D3; } #vcbkhkbzey .gt_striped { background-color: rgba(128, 128, 128, 0.05); } #vcbkhkbzey .gt_table_body { border-top-style: solid; border-top-width: 2px; border-top-color: #D3D3D3; border-bottom-style: solid; border-bottom-width: 2px; border-bottom-color: #D3D3D3; } #vcbkhkbzey .gt_footnotes { color: #333333; background-color: #FFFFFF; border-bottom-style: none; border-bottom-width: 2px; border-bottom-color: #D3D3D3; border-left-style: none; border-left-width: 2px; border-left-color: #D3D3D3; border-right-style: none; border-right-width: 2px; border-right-color: #D3D3D3; } #vcbkhkbzey .gt_footnote { margin: 0px; font-size: 90%; padding: 4px; } #vcbkhkbzey .gt_sourcenotes { color: #333333; background-color: #FFFFFF; border-bottom-style: none; border-bottom-width: 2px; border-bottom-color: #D3D3D3; border-left-style: none; border-left-width: 2px; border-left-color: #D3D3D3; border-right-style: none; border-right-width: 2px; border-right-color: #D3D3D3; } #vcbkhkbzey .gt_sourcenote { font-size: 90%; padding: 4px; } #vcbkhkbzey .gt_left { text-align: left; } #vcbkhkbzey .gt_center { text-align: center; } #vcbkhkbzey .gt_right { text-align: right; font-variant-numeric: tabular-nums; } #vcbkhkbzey .gt_font_normal { font-weight: normal; } #vcbkhkbzey .gt_font_bold { font-weight: bold; } #vcbkhkbzey .gt_font_italic { font-style: italic; } #vcbkhkbzey .gt_super { font-size: 65%; } #vcbkhkbzey .gt_footnote_marks { font-style: italic; font-size: 65%; } </style> <div id="vcbkhkbzey" style="overflow-x:auto;overflow-y:auto;width:auto;height:auto;"><table class="gt_table"> <thead class="gt_col_headings"> <tr> <th class="gt_col_heading gt_columns_bottom_border gt_left" rowspan="1" colspan="1" style="font-size: 21px;">name</th> <th class="gt_col_heading gt_columns_bottom_border gt_left" rowspan="1" colspan="1" style="font-size: 21px;">style</th> <th class="gt_col_heading gt_columns_bottom_border gt_right" rowspan="1" colspan="1" style="font-size: 21px;">alc_by_volume</th> <th class="gt_col_heading gt_columns_bottom_border gt_left" rowspan="1" colspan="1" style="font-size: 21px;">price_range</th> <th class="gt_col_heading gt_columns_bottom_border gt_left" rowspan="1" colspan="1" style="font-size: 21px;">country</th> <th class="gt_col_heading gt_columns_bottom_border gt_right" rowspan="1" colspan="1" style="font-size: 21px;">star_rating</th> </tr> </thead> <tbody class="gt_table_body"> <tr><td class="gt_row gt_left" style="font-size: 21px;">County</td> <td class="gt_row gt_left" style="font-size: 21px;">Lager</td> <td class="gt_row gt_right" style="font-size: 21px;">4.2</td> <td class="gt_row gt_left" style="font-size: 21px;">low</td> <td class="gt_row gt_left" style="font-size: 21px;">USA</td> <td class="gt_row gt_right" style="font-size: 21px;">3.40</td></tr> <tr><td class="gt_row gt_left" style="font-size: 21px;">Mountain Special</td> <td class="gt_row gt_left" style="font-size: 21px;">Pale Ale</td> <td class="gt_row gt_right" style="font-size: 21px;">4.1</td> <td class="gt_row gt_left" style="font-size: 21px;">low</td> <td class="gt_row gt_left" style="font-size: 21px;">USA</td> <td class="gt_row gt_right" style="font-size: 21px;">2.70</td></tr> <tr><td class="gt_row gt_left" style="font-size: 21px;">Lagartos</td> <td class="gt_row gt_left" style="font-size: 21px;">Lager</td> <td class="gt_row gt_right" style="font-size: 21px;">5.1</td> <td class="gt_row gt_left" style="font-size: 21px;">mid</td> <td class="gt_row gt_left" style="font-size: 21px;">Mexico</td> <td class="gt_row gt_right" style="font-size: 21px;">4.10</td></tr> <tr><td class="gt_row gt_left" style="font-size: 21px;">Jealous Hobo</td> <td class="gt_row gt_left" style="font-size: 21px;">Porter</td> <td class="gt_row gt_right" style="font-size: 21px;">6.5</td> <td class="gt_row gt_left" style="font-size: 21px;">mid</td> <td class="gt_row gt_left" style="font-size: 21px;">USA</td> <td class="gt_row gt_right" style="font-size: 21px;">3.36</td></tr> <tr><td class="gt_row gt_left" style="font-size: 21px;">Strasse</td> <td class="gt_row gt_left" style="font-size: 21px;">Stout</td> <td class="gt_row gt_right" style="font-size: 21px;">8.3</td> <td class="gt_row gt_left" style="font-size: 21px;">high</td> <td class="gt_row gt_left" style="font-size: 21px;">Germany</td> <td class="gt_row gt_right" style="font-size: 21px;">4.89</td></tr> <tr><td class="gt_row gt_left" style="font-size: 21px;">Arbre</td> <td class="gt_row gt_left" style="font-size: 21px;">Pale Ale</td> <td class="gt_row gt_right" style="font-size: 21px;">3.4</td> <td class="gt_row gt_left" style="font-size: 21px;">low</td> <td class="gt_row gt_left" style="font-size: 21px;">Belgium</td> <td class="gt_row gt_right" style="font-size: 21px;">4.01</td></tr> <tr><td class="gt_row gt_left" style="font-size: 21px;">K1997</td> <td class="gt_row gt_left" style="font-size: 21px;">Lager</td> <td class="gt_row gt_right" style="font-size: 21px;">4.0</td> <td class="gt_row gt_left" style="font-size: 21px;">high</td> <td class="gt_row gt_left" style="font-size: 21px;">Japan</td> <td class="gt_row gt_right" style="font-size: 21px;">3.88</td></tr> <tr><td class="gt_row gt_left" style="font-size: 21px;">Lagartos Cero</td> <td class="gt_row gt_left" style="font-size: 21px;">Lager</td> <td class="gt_row gt_right" style="font-size: 21px;">0.0</td> <td class="gt_row gt_left" style="font-size: 21px;">mid</td> <td class="gt_row gt_left" style="font-size: 21px;">Mexico</td> <td class="gt_row gt_right" style="font-size: 21px;">2.00</td></tr> <tr><td class="gt_row gt_left" style="font-size: 21px;">Airball</td> <td class="gt_row gt_left" style="font-size: 21px;">Irish Ale</td> <td class="gt_row gt_right" style="font-size: 21px;">6.0</td> <td class="gt_row gt_left" style="font-size: 21px;">high</td> <td class="gt_row gt_left" style="font-size: 21px;">USA</td> <td class="gt_row gt_right" style="font-size: 21px;">3.64</td></tr> <tr><td class="gt_row gt_left" style="font-size: 21px;">Clean Tackle</td> <td class="gt_row gt_left" style="font-size: 21px;">Stout</td> <td class="gt_row gt_right" style="font-size: 21px;">7.2</td> <td class="gt_row gt_left" style="font-size: 21px;">mid</td> <td class="gt_row gt_left" style="font-size: 21px;">England</td> <td class="gt_row gt_right" style="font-size: 21px;">4.20</td></tr> </tbody> </table></div> ] .panel[.panel-name[Data setup] ```r beers <- tibble::tribble( ~name, ~style, ~alc_by_volume, ~price_range, ~country, ~star_rating, "County", "Lager", 4.2, "low", "USA", 3.4, "Mountain Special", "Pale Ale", 4.1, "low", "USA", 2.7, "Lagartos", "Lager", 5.1, "mid", "Mexico", 4.1, "Jealous Hobo", "Porter", 6.5, "mid", "USA", 3.36, "Strasse", "Stout", 8.3, "high", "Germany", 4.89, "Arbre", "Pale Ale", 3.4, "low", "Belgium", 4.01, "K1997", "Lager", 4, "high", "Japan", 3.88, "Lagartos Cero", "Lager", 0, "mid", "Mexico", 2, "Airball", "Irish Ale", 6, "high", "USA", 3.64, "Clean Tackle", "Stout", 7.2, "mid", "England", 4.2 ) ``` ] ] ??? These data here describe some fictional beers and their properties. --- .left-column[ .large.b[`select()`] .large[Subset variables] - by name - by position - by type ] .right-column[ ```r beers %>% select(name, style, star_rating) ``` 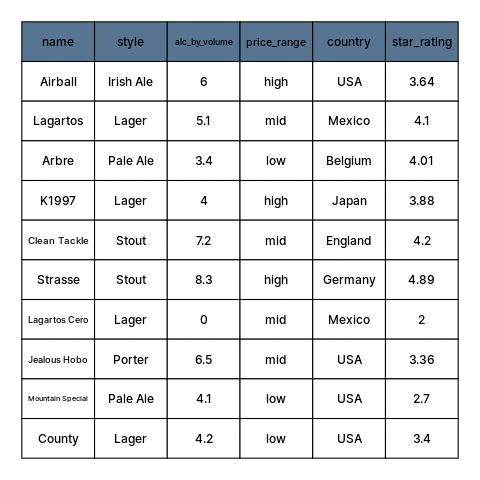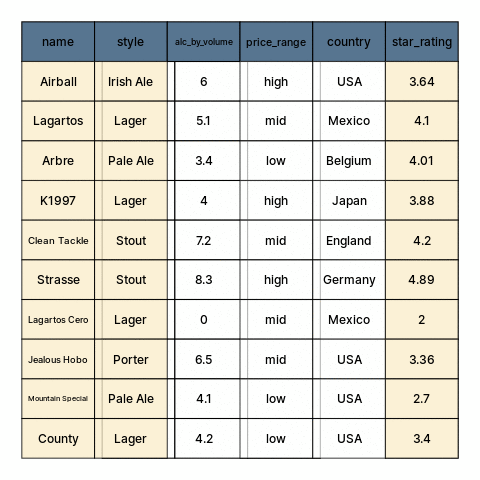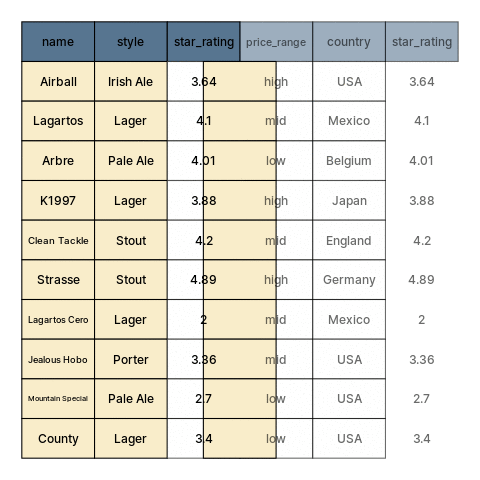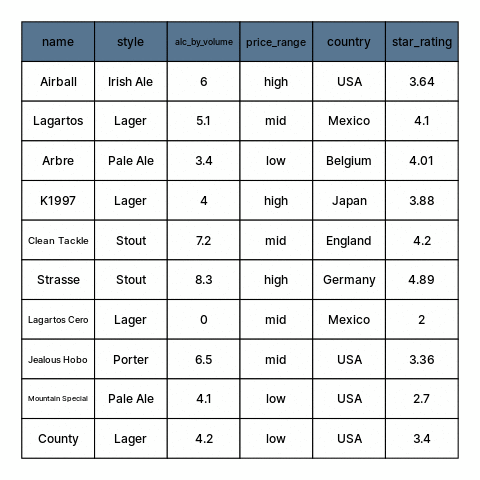 ] ??? The first thing we might want to try is to subset some variables from the original table. For this we use the select function. Select helps us subset variables, which we can specify by their name, position on the data from left to right, or even by type. In the example, we’re selecting three variables from the beers object, and we’re doing that with a pipe and by passing their names to the select function. The beers object is passed by the pipe as the first argument of the select function, and the other arguments are the names of the variables we want to keep, which are name, style, and star rating. --- .left-column[ .large.b[`mutate()`] .large[Create and modify columns] - create new - overwrite ] .right-column[ ```r beers %>% select(name, style, star_rating) %>% mutate(stars = round(star_rating, digits = 0)) ```  ] ??? Next up, is creating new columns of modifying existing ones. We can do this with the mutate function. The syntax for creating new columns is quite flexible, but typically we need to specify its name and what will be in this new column. Lets see whats happening in the example. To the transformation from before, in which we selected some variables from the beers object, we are adding an additional step using a pipe. With mutate, we’re creating a new variable called stars, which is itself the result of rounding the values of an existing column. When we use mutate, we create new columns, but if the name of the new column is the same as an existing one, it will be overwritten. --- .left-column[ .large.b[`filter()`] .large[subset rows] - keep rows that meet a condition ] .right-column[ ```r beers %>% select(name, style, star_rating) %>% mutate(stars = round(star_rating, digits = 0)) %>% filter(stars > 3) ```  ] ??? Finally, we can also work on rows. To subset rows that meet a condition, we use the filter function. Again, we’re using a pipe to add a new step to the sequence from before. With the new variable stars, we want to keep only the rows in which the values of stars are greater than 3. Notice how with the pipes we can keep track of the different steps, and that we didn’t have to work with intermediate objects for the results from each step. This is how we will work throughout this course. --- class: my-turn ## My Turn ??? Now, its my turn to use pipes and functions from dplyr to transform a dataset. -- - Load the mammal sleep data bundled with `ggplot2` -- - Select the "name" and "conservation" columns, as well as all numeric variables -- - Create a new column that contains the values of _'awake'_ multiplied by the values in _'brainwt'_ -- - Filter the data to keep only rows with _'sleep_total'_ > 4 -- > I'll use pipes to chain the operations --- class: inverse ## Your Turn -- - Load the mammal sleep data bundled with `ggplot2` -- - Select "name" and "conservation" columns and those that include the string _'sleep'_ in their name -- - Create a new column that contains the values of _'sleep_total'_ multiplied by 3 -- - Filter the data to remove domesticated mammals -- > Use pipes to chain the operations! --- class: center, middle, dk-section-title background-image:url("images/emmanuel-acua-43Tvb651fxE-unsplash.jpg") background-size: cover # Working with Columns with `across()` ??? In all the previous examples in which we selected or modified columns, we were working on them either one at a time or were having to name each one. --- ## Operating on multiple columns ###.center.b.large.orange[`across()`] -- `filter` or `mutate` multiple columns ??? Thanks to some recent updates to dplyr, we can operate on multiple columns at once using the across function. -- - By position, name, or type ??? We can specify many columns at once by position, name, or type. I don’t recommend using positions, because these are likely to change and less meaningful. -- - Compound selections of columns (e.g. _numeric columns that contain "mass" in their name_) ??? More importantly, we can use across to create compound selections of columns. For example, selecting or mutating all columns of type numeric but only if the contain a specific string in their name. --- ## .b.orange[`across()`] arguments: ??? Across has a relatively straight forward syntax. -- .large.b[`.cols`] Columns to transform ??? Cols is how we specify the columns we want to transform -- .large.b[`.fns`] Function to apply to each of the selected columns ??? and fns is the function to apply to each of the columns -- > Expects a function, so function name and arguments can be passed separately -- ```r beers %>% mutate(across(c(alc_by_volume, star_rating), round, digits = 3)) ``` ??? If you recall the beers data from earlier, we can use across to round all the numeric variables at the same time, the cols argument is a vector with the names of the two columns we’re interested in, and after that the function that we wish to apply and any additional arguments. Here, its the round function, with the digits argument passed on as well. Notice we used just the function name without any brackets. --- ## Useful column selection helpers ??? To specify sets of columns, there are a number of useful helper functions that we can use alongside or instead of the across function. -- .ba.ph4[ .b.orange[`everything()`] Match all variables ] </br> ```r beers %>% mutate(across(everything(), as.character)) ``` ??? First up is everything, which matches all variables. In the example, we’re transforming every variable in the beers object to character. --- ### Useful column selection helpers (continued) .ba.ph4[ .b.orange[`!`] Take the complement of a set of variables (negate a selection) ] </br> ```r beers %>% select(!c(style,country)) ``` ??? We can also use the exclamation mark to take the complement of a set of variables. In the example, the question mark negates the set of variables that are specified by name, so we get everything except style and country. --- ### Useful column selection helpers (continued) .ba.ph4.mt1.mb1[ .b.orange[`where()`] Selects variables for which a predicate function returns `TRUE` > e.g. subset or transform all numeric variables, determined by `is.numeric()`] ```r beers %>% mutate(across(where(is.numeric), log2)) ``` ??? Where is somewhat more elaborate, but it works together with any other function that returns TRUE of FALSE, and then select or mutates only those variables for which the condition is TRUE. For example, we want to apply a base two logarithm to any variables of type numeric in our beers object. We use the is.numeric function to check all the variables, and then the log2 function will only be applied to variables that meet the condition. --- ### Useful column selection helpers (continued) .ba.ph4[ .b.orange[`matches()`] Match variable names with a regular expression > e.g. drop variables with numbers in their name ] ```r beers %>% mutate(across(matches("rating"), log2)) ``` ??? Matches is good for working with variable names and working only on those that contain a match for our pattern of interest. This is can be very useful, and we will see the power of regular expressions in a separate lesson. In the example code, were matching any variables in which the name contains the word rating, and applying a base two log transformation. --- ### Useful column selection helpers (continued) .ba.ph4[ .b.orange[`:`] Select a range of consecutive variables ] <br> ```r beers %>% select(style:country) ``` ??? The colon, when used as a selection helper, matches a range of variables that are next to each other in the data. In the example, we’ll be keeping style, country, and every other variable between these two. --- ### Useful column selection helpers (continued) .ba.ph4[ .b.orange[`-`] Exclude variables (return all elements except the one preceded by the subtraction operator) ] ```r beers %>% select(-country) ``` ??? Finally, we can use the minus sign to exclude variables from a selection. In the example, we exclude country and keep everything else. --- `beers` <style>html { font-family: -apple-system, BlinkMacSystemFont, 'Segoe UI', Roboto, Oxygen, Ubuntu, Cantarell, 'Helvetica Neue', 'Fira Sans', 'Droid Sans', Arial, sans-serif; } #likyegqoso .gt_table { display: table; border-collapse: collapse; margin-left: auto; margin-right: auto; color: #333333; font-size: 16px; font-weight: normal; font-style: normal; background-color: #f4f4f9; width: auto; border-top-style: solid; border-top-width: 2px; border-top-color: #A8A8A8; border-right-style: none; border-right-width: 2px; border-right-color: #D3D3D3; border-bottom-style: solid; border-bottom-width: 2px; border-bottom-color: #A8A8A8; border-left-style: none; border-left-width: 2px; border-left-color: #D3D3D3; } #likyegqoso .gt_heading { background-color: #f4f4f9; text-align: center; border-bottom-color: #f4f4f9; border-left-style: none; border-left-width: 1px; border-left-color: #D3D3D3; border-right-style: none; border-right-width: 1px; border-right-color: #D3D3D3; } #likyegqoso .gt_title { color: #333333; font-size: 125%; font-weight: initial; padding-top: 4px; padding-bottom: 4px; border-bottom-color: #f4f4f9; border-bottom-width: 0; } #likyegqoso .gt_subtitle { color: #333333; font-size: 85%; font-weight: initial; padding-top: 0; padding-bottom: 4px; border-top-color: #f4f4f9; border-top-width: 0; } #likyegqoso .gt_bottom_border { border-bottom-style: solid; border-bottom-width: 2px; border-bottom-color: #D3D3D3; } #likyegqoso .gt_col_headings { border-top-style: solid; border-top-width: 2px; border-top-color: #D3D3D3; border-bottom-style: solid; border-bottom-width: 2px; border-bottom-color: #D3D3D3; border-left-style: none; border-left-width: 1px; border-left-color: #D3D3D3; border-right-style: none; border-right-width: 1px; border-right-color: #D3D3D3; } #likyegqoso .gt_col_heading { color: #333333; background-color: #f4f4f9; font-size: 100%; font-weight: normal; text-transform: inherit; border-left-style: none; border-left-width: 1px; border-left-color: #D3D3D3; border-right-style: none; border-right-width: 1px; border-right-color: #D3D3D3; vertical-align: bottom; padding-top: 5px; padding-bottom: 6px; padding-left: 5px; padding-right: 5px; overflow-x: hidden; } #likyegqoso .gt_column_spanner_outer { color: #333333; background-color: #f4f4f9; font-size: 100%; font-weight: normal; text-transform: inherit; padding-top: 0; padding-bottom: 0; padding-left: 4px; padding-right: 4px; } #likyegqoso .gt_column_spanner_outer:first-child { padding-left: 0; } #likyegqoso .gt_column_spanner_outer:last-child { padding-right: 0; } #likyegqoso .gt_column_spanner { border-bottom-style: solid; border-bottom-width: 2px; border-bottom-color: #D3D3D3; vertical-align: bottom; padding-top: 5px; padding-bottom: 6px; overflow-x: hidden; display: inline-block; width: 100%; } #likyegqoso .gt_group_heading { padding: 8px; color: #333333; background-color: #f4f4f9; font-size: 100%; font-weight: initial; text-transform: inherit; border-top-style: solid; border-top-width: 2px; border-top-color: #D3D3D3; border-bottom-style: solid; border-bottom-width: 2px; border-bottom-color: #D3D3D3; border-left-style: none; border-left-width: 1px; border-left-color: #D3D3D3; border-right-style: none; border-right-width: 1px; border-right-color: #D3D3D3; vertical-align: middle; } #likyegqoso .gt_empty_group_heading { padding: 0.5px; color: #333333; background-color: #f4f4f9; font-size: 100%; font-weight: initial; border-top-style: solid; border-top-width: 2px; border-top-color: #D3D3D3; border-bottom-style: solid; border-bottom-width: 2px; border-bottom-color: #D3D3D3; vertical-align: middle; } #likyegqoso .gt_from_md > :first-child { margin-top: 0; } #likyegqoso .gt_from_md > :last-child { margin-bottom: 0; } #likyegqoso .gt_row { padding-top: 8px; padding-bottom: 8px; padding-left: 5px; padding-right: 5px; margin: 10px; border-top-style: solid; border-top-width: 1px; border-top-color: #D3D3D3; border-left-style: none; border-left-width: 1px; border-left-color: #D3D3D3; border-right-style: none; border-right-width: 1px; border-right-color: #D3D3D3; vertical-align: middle; overflow-x: hidden; } #likyegqoso .gt_stub { color: #333333; background-color: #f4f4f9; font-size: 100%; font-weight: initial; text-transform: inherit; border-right-style: solid; border-right-width: 2px; border-right-color: #D3D3D3; padding-left: 12px; } #likyegqoso .gt_summary_row { color: #333333; background-color: #f4f4f9; text-transform: inherit; padding-top: 8px; padding-bottom: 8px; padding-left: 5px; padding-right: 5px; } #likyegqoso .gt_first_summary_row { padding-top: 8px; padding-bottom: 8px; padding-left: 5px; padding-right: 5px; border-top-style: solid; border-top-width: 2px; border-top-color: #D3D3D3; } #likyegqoso .gt_grand_summary_row { color: #333333; background-color: #f4f4f9; text-transform: inherit; padding-top: 8px; padding-bottom: 8px; padding-left: 5px; padding-right: 5px; } #likyegqoso .gt_first_grand_summary_row { padding-top: 8px; padding-bottom: 8px; padding-left: 5px; padding-right: 5px; border-top-style: double; border-top-width: 6px; border-top-color: #D3D3D3; } #likyegqoso .gt_striped { background-color: rgba(128, 128, 128, 0.05); } #likyegqoso .gt_table_body { border-top-style: solid; border-top-width: 2px; border-top-color: #D3D3D3; border-bottom-style: solid; border-bottom-width: 2px; border-bottom-color: #D3D3D3; } #likyegqoso .gt_footnotes { color: #333333; background-color: #f4f4f9; border-bottom-style: none; border-bottom-width: 2px; border-bottom-color: #D3D3D3; border-left-style: none; border-left-width: 2px; border-left-color: #D3D3D3; border-right-style: none; border-right-width: 2px; border-right-color: #D3D3D3; } #likyegqoso .gt_footnote { margin: 0px; font-size: 90%; padding: 4px; } #likyegqoso .gt_sourcenotes { color: #333333; background-color: #f4f4f9; border-bottom-style: none; border-bottom-width: 2px; border-bottom-color: #D3D3D3; border-left-style: none; border-left-width: 2px; border-left-color: #D3D3D3; border-right-style: none; border-right-width: 2px; border-right-color: #D3D3D3; } #likyegqoso .gt_sourcenote { font-size: 90%; padding: 4px; } #likyegqoso .gt_left { text-align: left; } #likyegqoso .gt_center { text-align: center; } #likyegqoso .gt_right { text-align: right; font-variant-numeric: tabular-nums; } #likyegqoso .gt_font_normal { font-weight: normal; } #likyegqoso .gt_font_bold { font-weight: bold; } #likyegqoso .gt_font_italic { font-style: italic; } #likyegqoso .gt_super { font-size: 65%; } #likyegqoso .gt_footnote_marks { font-style: italic; font-size: 65%; } </style> <div id="likyegqoso" style="overflow-x:auto;overflow-y:auto;width:auto;height:auto;"><table class="gt_table"> <thead class="gt_col_headings"> <tr> <th class="gt_col_heading gt_columns_bottom_border gt_left" rowspan="1" colspan="1" style="font-size: 21px;">name</th> <th class="gt_col_heading gt_columns_bottom_border gt_left" rowspan="1" colspan="1" style="font-size: 21px;">style</th> <th class="gt_col_heading gt_columns_bottom_border gt_right" rowspan="1" colspan="1" style="font-size: 21px;">alc_by_volume</th> <th class="gt_col_heading gt_columns_bottom_border gt_left" rowspan="1" colspan="1" style="font-size: 21px;">price_range</th> <th class="gt_col_heading gt_columns_bottom_border gt_left" rowspan="1" colspan="1" style="font-size: 21px;">country</th> <th class="gt_col_heading gt_columns_bottom_border gt_right" rowspan="1" colspan="1" style="font-size: 21px;">star_rating</th> </tr> </thead> <tbody class="gt_table_body"> <tr><td class="gt_row gt_left" style="font-size: 21px;">County</td> <td class="gt_row gt_left" style="font-size: 21px;">Lager</td> <td class="gt_row gt_right" style="font-size: 21px;">4.2</td> <td class="gt_row gt_left" style="font-size: 21px;">low</td> <td class="gt_row gt_left" style="font-size: 21px;">USA</td> <td class="gt_row gt_right" style="font-size: 21px;">3.40</td></tr> <tr><td class="gt_row gt_left" style="font-size: 21px;">Mountain Special</td> <td class="gt_row gt_left" style="font-size: 21px;">Pale Ale</td> <td class="gt_row gt_right" style="font-size: 21px;">4.1</td> <td class="gt_row gt_left" style="font-size: 21px;">low</td> <td class="gt_row gt_left" style="font-size: 21px;">USA</td> <td class="gt_row gt_right" style="font-size: 21px;">2.70</td></tr> <tr><td class="gt_row gt_left" style="font-size: 21px;">Lagartos</td> <td class="gt_row gt_left" style="font-size: 21px;">Lager</td> <td class="gt_row gt_right" style="font-size: 21px;">5.1</td> <td class="gt_row gt_left" style="font-size: 21px;">mid</td> <td class="gt_row gt_left" style="font-size: 21px;">Mexico</td> <td class="gt_row gt_right" style="font-size: 21px;">4.10</td></tr> <tr><td class="gt_row gt_left" style="font-size: 21px;">Jealous Hobo</td> <td class="gt_row gt_left" style="font-size: 21px;">Porter</td> <td class="gt_row gt_right" style="font-size: 21px;">6.5</td> <td class="gt_row gt_left" style="font-size: 21px;">mid</td> <td class="gt_row gt_left" style="font-size: 21px;">USA</td> <td class="gt_row gt_right" style="font-size: 21px;">3.36</td></tr> <tr><td class="gt_row gt_left" style="font-size: 21px;">Strasse</td> <td class="gt_row gt_left" style="font-size: 21px;">Stout</td> <td class="gt_row gt_right" style="font-size: 21px;">8.3</td> <td class="gt_row gt_left" style="font-size: 21px;">high</td> <td class="gt_row gt_left" style="font-size: 21px;">Germany</td> <td class="gt_row gt_right" style="font-size: 21px;">4.89</td></tr> <tr><td class="gt_row gt_left" style="font-size: 21px;">Arbre</td> <td class="gt_row gt_left" style="font-size: 21px;">Pale Ale</td> <td class="gt_row gt_right" style="font-size: 21px;">3.4</td> <td class="gt_row gt_left" style="font-size: 21px;">low</td> <td class="gt_row gt_left" style="font-size: 21px;">Belgium</td> <td class="gt_row gt_right" style="font-size: 21px;">4.01</td></tr> <tr><td class="gt_row gt_left" style="font-size: 21px;">K1997</td> <td class="gt_row gt_left" style="font-size: 21px;">Lager</td> <td class="gt_row gt_right" style="font-size: 21px;">4.0</td> <td class="gt_row gt_left" style="font-size: 21px;">high</td> <td class="gt_row gt_left" style="font-size: 21px;">Japan</td> <td class="gt_row gt_right" style="font-size: 21px;">3.88</td></tr> <tr><td class="gt_row gt_left" style="font-size: 21px;">Lagartos Cero</td> <td class="gt_row gt_left" style="font-size: 21px;">Lager</td> <td class="gt_row gt_right" style="font-size: 21px;">0.0</td> <td class="gt_row gt_left" style="font-size: 21px;">mid</td> <td class="gt_row gt_left" style="font-size: 21px;">Mexico</td> <td class="gt_row gt_right" style="font-size: 21px;">2.00</td></tr> <tr><td class="gt_row gt_left" style="font-size: 21px;">Airball</td> <td class="gt_row gt_left" style="font-size: 21px;">Irish Ale</td> <td class="gt_row gt_right" style="font-size: 21px;">6.0</td> <td class="gt_row gt_left" style="font-size: 21px;">high</td> <td class="gt_row gt_left" style="font-size: 21px;">USA</td> <td class="gt_row gt_right" style="font-size: 21px;">3.64</td></tr> <tr><td class="gt_row gt_left" style="font-size: 21px;">Clean Tackle</td> <td class="gt_row gt_left" style="font-size: 21px;">Stout</td> <td class="gt_row gt_right" style="font-size: 21px;">7.2</td> <td class="gt_row gt_left" style="font-size: 21px;">mid</td> <td class="gt_row gt_left" style="font-size: 21px;">England</td> <td class="gt_row gt_right" style="font-size: 21px;">4.20</td></tr> </tbody> </table></div> ??? Lets have another look at the beers data, and apply some transformations to many columns at once. Just looking at this table, we can see two numeric variables: alcohol by volume, and star rating. Just for demonstration, lets use the mutate function to apply a numeric transformation to these two columns. --- ### Square root (.orange.b[`sqrt()`]) of `alc_by_volume` and `star_rating` ```r beers %>% mutate(across(c(alc_by_volume,star_rating), sqrt)) ``` ``` ## # A tibble: 10 x 6 ## name style alc_by_volume price_range country star_rating ## <chr> <chr> <dbl> <chr> <chr> <dbl> ## 1 County Lager 2.05 low USA 1.84 ## 2 Mountain Special Pale Ale 2.02 low USA 1.64 ## 3 Lagartos Lager 2.26 mid Mexico 2.02 ## 4 Jealous Hobo Porter 2.55 mid USA 1.83 ## 5 Strasse Stout 2.88 high Germany 2.21 ## 6 Arbre Pale Ale 1.84 low Belgium 2.00 ## 7 K1997 Lager 2 high Japan 1.97 ## 8 Lagartos Cero Lager 0 mid Mexico 1.41 ## 9 Airball Irish Ale 2.45 high USA 1.91 ## 10 Clean Tackle Stout 2.68 mid England 2.05 ``` ??? To apply the square root function to these two columns, one option would be to refer to them by name inside across. --- ### Square root (.orange.b[`sqrt()`]) of all numeric variables ```r beers %>% mutate(across(where(is.numeric), sqrt)) ``` ``` ## # A tibble: 10 x 6 ## name style alc_by_volume price_range country star_rating ## <chr> <chr> <dbl> <chr> <chr> <dbl> ## 1 County Lager 2.05 low USA 1.84 ## 2 Mountain Special Pale Ale 2.02 low USA 1.64 ## 3 Lagartos Lager 2.26 mid Mexico 2.02 ## 4 Jealous Hobo Porter 2.55 mid USA 1.83 ## 5 Strasse Stout 2.88 high Germany 2.21 ## 6 Arbre Pale Ale 1.84 low Belgium 2.00 ## 7 K1997 Lager 2 high Japan 1.97 ## 8 Lagartos Cero Lager 0 mid Mexico 1.41 ## 9 Airball Irish Ale 2.45 high USA 1.91 ## 10 Clean Tackle Stout 2.68 mid England 2.05 ``` ??? Or, we can also use the where function and is.numeric to simplify our code and apply the square root function to every numeric variable. The results are both the same. --- class: my-turn ## My Turn -- - Load the _midwest_ data bundled with `ggplot2` -- - Keep only rows for Wisconsin (WI) -- - Subset the 'county' column and all columns that match the string '_perc_' (hint: use a selection helper) -- - log-transform all numeric variables -- - Append the suffix "_check" to every value --- class: inverse ## Your Turn -- - Load the _midwest_ data bundled with `ggplot2` -- - Keep only rows for Ohio (OH) -- - Subset the 'county' column and all columns that match the string '_pop_' (hint: use a selection helper) -- - Square-root transform all numeric variables --- class: center, middle, dk-section-title background-image:url("images/1024px-Espacio_Escultórico.jpg") background-size: cover # Pivoting Data ??? In the lesson on data organization, we went over tidy data principles, and glanced over long and wide structures for data. Now, we can go ahead and use R to reshape and pivot our data between these different structures. --- .pull-left[ ## Wide Data Human-readable Easier entry Easier interactive editing ] .pull.right[ ## Long Data Often meets Tidy Data principles Analysis-ready Maps well to plot axes and aesthetics ]  ??? As the names suggest, data can take either wide and long structures. The difference between these two structures goes beyond just having different proportions of rows and columns. This goes back to tidy data principles. Wide data is what we see often and what we use for data entry or reporting. This structure is readable, concise, and easy to edit. On the other hand, long data is more likely to be tidy and more ready for use in our workflows. The fundamental difference lies with how variables and observations relate to rows and columns. The good news is that we can transform the data between structures. --- # Reshaping data with 📦 .b.rrured[`tidyr`] ??? To reshape data from wide to long and vice versa, we’ll use functions from the tidyr package, which, if you remember, is part of the tidyverse. The names of the relevant functions are also quite easy to remember. -- .large[wide to long: .b.orange[`pivot_longer()`]] ```r pivot_longer(data, cols, names_to, values_to) ``` ??? To go from wide to long, we use pivot_longer -- .large[long to wide: .b.orange[`pivot_wider()`]] ```r pivot_longer(data, cols, names_from, values_from) ``` ??? To go from long to wide, we use pivot_wider --- .panelset[ .panel[.panel-name[critters] <style>html { font-family: -apple-system, BlinkMacSystemFont, 'Segoe UI', Roboto, Oxygen, Ubuntu, Cantarell, 'Helvetica Neue', 'Fira Sans', 'Droid Sans', Arial, sans-serif; } #jiplyrgdqm .gt_table { display: table; border-collapse: collapse; margin-left: auto; margin-right: auto; color: #333333; font-size: 16px; font-weight: normal; font-style: normal; background-color: #f4f4f9; width: auto; border-top-style: solid; border-top-width: 2px; border-top-color: #A8A8A8; border-right-style: none; border-right-width: 2px; border-right-color: #D3D3D3; border-bottom-style: solid; border-bottom-width: 2px; border-bottom-color: #A8A8A8; border-left-style: none; border-left-width: 2px; border-left-color: #D3D3D3; } #jiplyrgdqm .gt_heading { background-color: #f4f4f9; text-align: center; border-bottom-color: #f4f4f9; border-left-style: none; border-left-width: 1px; border-left-color: #D3D3D3; border-right-style: none; border-right-width: 1px; border-right-color: #D3D3D3; } #jiplyrgdqm .gt_title { color: #333333; font-size: 125%; font-weight: initial; padding-top: 4px; padding-bottom: 4px; border-bottom-color: #f4f4f9; border-bottom-width: 0; } #jiplyrgdqm .gt_subtitle { color: #333333; font-size: 85%; font-weight: initial; padding-top: 0; padding-bottom: 4px; border-top-color: #f4f4f9; border-top-width: 0; } #jiplyrgdqm .gt_bottom_border { border-bottom-style: solid; border-bottom-width: 2px; border-bottom-color: #D3D3D3; } #jiplyrgdqm .gt_col_headings { border-top-style: solid; border-top-width: 2px; border-top-color: #D3D3D3; border-bottom-style: solid; border-bottom-width: 2px; border-bottom-color: #D3D3D3; border-left-style: none; border-left-width: 1px; border-left-color: #D3D3D3; border-right-style: none; border-right-width: 1px; border-right-color: #D3D3D3; } #jiplyrgdqm .gt_col_heading { color: #333333; background-color: #f4f4f9; font-size: 100%; font-weight: normal; text-transform: inherit; border-left-style: none; border-left-width: 1px; border-left-color: #D3D3D3; border-right-style: none; border-right-width: 1px; border-right-color: #D3D3D3; vertical-align: bottom; padding-top: 5px; padding-bottom: 6px; padding-left: 5px; padding-right: 5px; overflow-x: hidden; } #jiplyrgdqm .gt_column_spanner_outer { color: #333333; background-color: #f4f4f9; font-size: 100%; font-weight: normal; text-transform: inherit; padding-top: 0; padding-bottom: 0; padding-left: 4px; padding-right: 4px; } #jiplyrgdqm .gt_column_spanner_outer:first-child { padding-left: 0; } #jiplyrgdqm .gt_column_spanner_outer:last-child { padding-right: 0; } #jiplyrgdqm .gt_column_spanner { border-bottom-style: solid; border-bottom-width: 2px; border-bottom-color: #D3D3D3; vertical-align: bottom; padding-top: 5px; padding-bottom: 6px; overflow-x: hidden; display: inline-block; width: 100%; } #jiplyrgdqm .gt_group_heading { padding: 8px; color: #333333; background-color: #f4f4f9; font-size: 100%; font-weight: initial; text-transform: inherit; border-top-style: solid; border-top-width: 2px; border-top-color: #D3D3D3; border-bottom-style: solid; border-bottom-width: 2px; border-bottom-color: #D3D3D3; border-left-style: none; border-left-width: 1px; border-left-color: #D3D3D3; border-right-style: none; border-right-width: 1px; border-right-color: #D3D3D3; vertical-align: middle; } #jiplyrgdqm .gt_empty_group_heading { padding: 0.5px; color: #333333; background-color: #f4f4f9; font-size: 100%; font-weight: initial; border-top-style: solid; border-top-width: 2px; border-top-color: #D3D3D3; border-bottom-style: solid; border-bottom-width: 2px; border-bottom-color: #D3D3D3; vertical-align: middle; } #jiplyrgdqm .gt_from_md > :first-child { margin-top: 0; } #jiplyrgdqm .gt_from_md > :last-child { margin-bottom: 0; } #jiplyrgdqm .gt_row { padding-top: 8px; padding-bottom: 8px; padding-left: 5px; padding-right: 5px; margin: 10px; border-top-style: solid; border-top-width: 1px; border-top-color: #D3D3D3; border-left-style: none; border-left-width: 1px; border-left-color: #D3D3D3; border-right-style: none; border-right-width: 1px; border-right-color: #D3D3D3; vertical-align: middle; overflow-x: hidden; } #jiplyrgdqm .gt_stub { color: #333333; background-color: #f4f4f9; font-size: 100%; font-weight: initial; text-transform: inherit; border-right-style: solid; border-right-width: 2px; border-right-color: #D3D3D3; padding-left: 12px; } #jiplyrgdqm .gt_summary_row { color: #333333; background-color: #f4f4f9; text-transform: inherit; padding-top: 8px; padding-bottom: 8px; padding-left: 5px; padding-right: 5px; } #jiplyrgdqm .gt_first_summary_row { padding-top: 8px; padding-bottom: 8px; padding-left: 5px; padding-right: 5px; border-top-style: solid; border-top-width: 2px; border-top-color: #D3D3D3; } #jiplyrgdqm .gt_grand_summary_row { color: #333333; background-color: #f4f4f9; text-transform: inherit; padding-top: 8px; padding-bottom: 8px; padding-left: 5px; padding-right: 5px; } #jiplyrgdqm .gt_first_grand_summary_row { padding-top: 8px; padding-bottom: 8px; padding-left: 5px; padding-right: 5px; border-top-style: double; border-top-width: 6px; border-top-color: #D3D3D3; } #jiplyrgdqm .gt_striped { background-color: rgba(128, 128, 128, 0.05); } #jiplyrgdqm .gt_table_body { border-top-style: solid; border-top-width: 2px; border-top-color: #D3D3D3; border-bottom-style: solid; border-bottom-width: 2px; border-bottom-color: #D3D3D3; } #jiplyrgdqm .gt_footnotes { color: #333333; background-color: #f4f4f9; border-bottom-style: none; border-bottom-width: 2px; border-bottom-color: #D3D3D3; border-left-style: none; border-left-width: 2px; border-left-color: #D3D3D3; border-right-style: none; border-right-width: 2px; border-right-color: #D3D3D3; } #jiplyrgdqm .gt_footnote { margin: 0px; font-size: 90%; padding: 4px; } #jiplyrgdqm .gt_sourcenotes { color: #333333; background-color: #f4f4f9; border-bottom-style: none; border-bottom-width: 2px; border-bottom-color: #D3D3D3; border-left-style: none; border-left-width: 2px; border-left-color: #D3D3D3; border-right-style: none; border-right-width: 2px; border-right-color: #D3D3D3; } #jiplyrgdqm .gt_sourcenote { font-size: 90%; padding: 4px; } #jiplyrgdqm .gt_left { text-align: left; } #jiplyrgdqm .gt_center { text-align: center; } #jiplyrgdqm .gt_right { text-align: right; font-variant-numeric: tabular-nums; } #jiplyrgdqm .gt_font_normal { font-weight: normal; } #jiplyrgdqm .gt_font_bold { font-weight: bold; } #jiplyrgdqm .gt_font_italic { font-style: italic; } #jiplyrgdqm .gt_super { font-size: 65%; } #jiplyrgdqm .gt_footnote_marks { font-style: italic; font-size: 65%; } </style> <div id="jiplyrgdqm" style="overflow-x:auto;overflow-y:auto;width:auto;height:auto;"><table class="gt_table"> <thead class="gt_col_headings"> <tr> <th class="gt_col_heading gt_columns_bottom_border gt_left" rowspan="1" colspan="1" style="font-size: 22px;">Habitat</th> <th class="gt_col_heading gt_columns_bottom_border gt_left" rowspan="1" colspan="1" style="font-size: 22px;">County</th> <th class="gt_col_heading gt_columns_bottom_border gt_right" rowspan="1" colspan="1" style="font-size: 22px;">Frogs</th> <th class="gt_col_heading gt_columns_bottom_border gt_right" rowspan="1" colspan="1" style="font-size: 22px;">Crayfish</th> <th class="gt_col_heading gt_columns_bottom_border gt_right" rowspan="1" colspan="1" style="font-size: 22px;">Owls</th> </tr> </thead> <tbody class="gt_table_body"> <tr><td class="gt_row gt_left" style="font-size: 22px;">Marsh</td> <td class="gt_row gt_left" style="font-size: 22px;">Clarkville</td> <td class="gt_row gt_right" style="font-size: 22px;">7</td> <td class="gt_row gt_right" style="font-size: 22px;">5</td> <td class="gt_row gt_right" style="font-size: 22px;">4</td></tr> <tr><td class="gt_row gt_left" style="font-size: 22px;">Urban</td> <td class="gt_row gt_left" style="font-size: 22px;">Smith</td> <td class="gt_row gt_right" style="font-size: 22px;">0</td> <td class="gt_row gt_right" style="font-size: 22px;">0</td> <td class="gt_row gt_right" style="font-size: 22px;">2</td></tr> <tr><td class="gt_row gt_left" style="font-size: 22px;">Wetland</td> <td class="gt_row gt_left" style="font-size: 22px;">Wilson</td> <td class="gt_row gt_right" style="font-size: 22px;">9</td> <td class="gt_row gt_right" style="font-size: 22px;">2</td> <td class="gt_row gt_right" style="font-size: 22px;">1</td></tr> <tr><td class="gt_row gt_left" style="font-size: 22px;">Forest</td> <td class="gt_row gt_left" style="font-size: 22px;">Smith</td> <td class="gt_row gt_right" style="font-size: 22px;">4</td> <td class="gt_row gt_right" style="font-size: 22px;">0</td> <td class="gt_row gt_right" style="font-size: 22px;">7</td></tr> </tbody> </table></div> ] .panel[.panel-name[Data setup] ```r critters <- tibble::tribble( ~Habitat, ~County, ~Frogs, ~Crayfish, ~Owls, "Marsh", "Clarkville", 7L, 5L, 4L, "Urban", "Smith", 0L, 0L, 2L, "Wetland", "Wilson", 9L, 2L, 1L, "Forest", "Smith", 4L, 0L, 7L ) ``` ] ] ??? Ill illustrate these transformations with this simple dataset that enumerates some wildlife sightings across different habitats and counties. Just looking at this table, we can see that it is wider than it is long. -- .center[Variables (attributes measured):] .fl.w-24[ Habitat ] .fl.w-25.tc[ County ] .fl.w-25.tc[ Critter Type ] .fl.w-25.tr[ Individuals Encountered ] ??? Going deeper into the structure of the data though, we will come to realize that this wide data is not tidy. Why? Because there are actually four attributes being measured, but their values do not correspond to columns. Specifically, the values for critter type and the number of individuals encountered are not in their own column. We can still interpret the information stored in these data, but for many purposes a long, tidy version might be preferable. Lets see how we can go about reshaping this critter data. --- # Wide to long ```r critters_long <- critters %>% pivot_longer( cols = c(Frogs, Crayfish, Owls), names_to = "critter_type", values_to = "individuals_encountered" ) ``` ??? For this, we use the pivot longer function. Notice again how we’re using pipes. Pivot longer has three main arguments that we need to specify. -- .b[`cols`] Which columns have data in their names ??? Cols are the columns that actually contain data in their names. In our example, these would be the columns for each of the little animals, which are actually levels of an attribute. -- **`names_to`** Name for the new column to be created from `cols` ??? Having recognized that there were data in column names which should be in its own column, we can give it a name. This is what the names to argument is for. In this case, lets call the new column critter underscore type -- **`values_to`** Name of the column to be created from the contents of the cells in **`cols`** ??? finally, the content of the columns we specified with the cols argument also needs to go into a separate variable, and it will need a name. In our example, this would be the number of individuals spotted, and we can use that as a name. --- ```r critters %>% pivot_longer(cols = c(Frogs, Crayfish, Owls), names_to = "critter_type", values_to = "individuals_encountered") ``` ``` ## # A tibble: 12 x 4 ## Habitat County critter_type individuals_encountered ## <chr> <chr> <chr> <int> ## 1 Marsh Clarkville Frogs 7 ## 2 Marsh Clarkville Crayfish 5 ## 3 Marsh Clarkville Owls 4 ## 4 Urban Smith Frogs 0 ## 5 Urban Smith Crayfish 0 ## 6 Urban Smith Owls 2 ## 7 Wetland Wilson Frogs 9 ## 8 Wetland Wilson Crayfish 2 ## 9 Wetland Wilson Owls 1 ## 10 Forest Smith Frogs 4 ## 11 Forest Smith Crayfish 0 ## 12 Forest Smith Owls 7 ``` ??? Lets see how the long table looks. The same information is now in a long, tidy version. Notice how the function handled all the combinations for us, and how there is more repetition with this structure. Now, each of the four measured attributes is in a column and each row is now an observation with a value for each variable. --- # Long to wide ```r critters_long %>% pivot_wider(names_from = critter_type, values_from = individuals_encountered) ``` ??? We can just as easily reshape the long version back to a wide version. For this, we use two main arguments. -- .b[`names_from`] Which columns' values will be used for new .b[column names] ??? First, we need to specify which column contains the values that need to be spread out across as many column names as there are unique values in the column. We do this with the names from argument. -- .b[`values_from`] Which column has the .b[cell values] for the new output columns ??? Then, we need to specify which column has the values that will populate the new columns. We use values from for this. --- ```r critters_long %>% pivot_wider(names_from = critter_type, values_from = individuals_encountered) ``` ``` ## # A tibble: 4 x 5 ## Habitat County Frogs Crayfish Owls ## <chr> <chr> <int> <int> <int> ## 1 Marsh Clarkville 7 5 4 ## 2 Urban Smith 0 0 2 ## 3 Wetland Wilson 9 2 1 ## 4 Forest Smith 4 0 7 ``` ??? With our example, the new “critter type” column has the names of the critters that we want as separate columns, and the “individuals encountered” column has the numeric values that will also be spread out into the new columns. When we run this code, we get our initial wide dataset back. --- class: center  ??? Maybe an animated version of this transformation can illustrate the process better. Look at how the same information can appear in different layouts. Each one has its advantages and disadvantages, but its important to know that its not an issue shifting between the two. --- class: my-turn # My Turn -- - .large[Load the dog ranks data ("dogranks_mine.csv")] -- - .large[Pivot the data (wide to long and back to wide)] --- class: inverse # Your Turn -- - .large[Load the dog ranks data ("dogranks_your.csv")] -- - .large[Pivot the data (wide to long and back to wide)] --- class: center, middle, dk-section-title background-image:url("images/drew-dau-dXxsLrlg9qY-unsplash.jpg") background-size: cover # Coalescing Columns ??? By now, we’ve gone over subsetting rows and columns, creating new columns, and reshaping entire datasets between wide and long structures. These tools are pretty much everything we need to restructure our data. Now, we’ll learn about two more functions from dplyr and tidyr that may come in handy during the restructuring stage of our data cleaning. First, we’ll learn about coalescing columns. Coalescing values sounds strange and complicated, but as we’ll see, this comes up often during data cleaning and the important thing is that we can avoid having to do this by hand. Coalescing columns is essentially lumping together two or more columns with data for the same variable, in which there may be missing values. We want to combine the data, keeping values from one column and filling any missing data with values from other columns. --- ## .b.orange[coalesce()] from 📦 .b.rrured[`dplyr`] ??? To do this, we use the coalesce function from dplyr. -- - Find and return the first non-NA value at each position for a set of vectors ??? Technically, coalesce finds and returns the first non NA value at each position for two or more vectors. Remember, columns in rectangular data are also vectors. -- - Returns a single vector with the non-NA values that appear first ??? When we coalesce a set of vectors, the function returns a single vector with all the non NA values, according to where they appear. --- ## .b.orange[coalesce()] from 📦 .b.rrured[`dplyr`] ```r x <- c(1, 2, NA, NA, 5) y <- c(NA, NA, 3, 4, 5) z <- c(1, 4, 3, 5, 3) ``` ??? Lets demonstrate this with some simple vectors. X Y and Z are numeric vectors of the same length and some have missing values. -- ```r coalesce(x, y, z) ``` ``` ## [1] 1 2 3 4 5 ``` ??? We want a single vector with as few Nas as possible. If we coalesce these three vectors in this particular order, the values from vector Y at the third and fourth positions fill in the missing values in vector X, and then vector Z is not used. The order of the vectors here is important. --- ## .b.orange[coalesce()] from 📦 .b.rrured[`dplyr`] ```r x <- c(1, 2, NA, NA, 5) y <- c(NA, NA, 3, 4, 5) z <- c(1, 4, 3, 5, 3) ``` ??? Lets do this again, but in a different order, this time z, x, and y. There are no missing values in vector Z so it is returned as is. -- ```r coalesce(z,x,y) ``` ``` ## [1] 1 4 3 5 3 ``` ??? Coalesce is a very useful function, but we need to be mindful of how we order and prioritize its inputs. Lets use coalesce in a more realistic scenario, to really illustrate its potential. --- .panelset[ .panel[.panel-name[sensors] <style>html { font-family: -apple-system, BlinkMacSystemFont, 'Segoe UI', Roboto, Oxygen, Ubuntu, Cantarell, 'Helvetica Neue', 'Fira Sans', 'Droid Sans', Arial, sans-serif; } #kfibdwimrn .gt_table { display: table; border-collapse: collapse; margin-left: auto; margin-right: auto; color: #333333; font-size: 16px; font-weight: normal; font-style: normal; background-color: #f4f4f9; width: auto; border-top-style: solid; border-top-width: 2px; border-top-color: #A8A8A8; border-right-style: none; border-right-width: 2px; border-right-color: #D3D3D3; border-bottom-style: solid; border-bottom-width: 2px; border-bottom-color: #A8A8A8; border-left-style: none; border-left-width: 2px; border-left-color: #D3D3D3; } #kfibdwimrn .gt_heading { background-color: #f4f4f9; text-align: center; border-bottom-color: #f4f4f9; border-left-style: none; border-left-width: 1px; border-left-color: #D3D3D3; border-right-style: none; border-right-width: 1px; border-right-color: #D3D3D3; } #kfibdwimrn .gt_title { color: #333333; font-size: 125%; font-weight: initial; padding-top: 4px; padding-bottom: 4px; border-bottom-color: #f4f4f9; border-bottom-width: 0; } #kfibdwimrn .gt_subtitle { color: #333333; font-size: 85%; font-weight: initial; padding-top: 0; padding-bottom: 4px; border-top-color: #f4f4f9; border-top-width: 0; } #kfibdwimrn .gt_bottom_border { border-bottom-style: solid; border-bottom-width: 2px; border-bottom-color: #D3D3D3; } #kfibdwimrn .gt_col_headings { border-top-style: solid; border-top-width: 2px; border-top-color: #D3D3D3; border-bottom-style: solid; border-bottom-width: 2px; border-bottom-color: #D3D3D3; border-left-style: none; border-left-width: 1px; border-left-color: #D3D3D3; border-right-style: none; border-right-width: 1px; border-right-color: #D3D3D3; } #kfibdwimrn .gt_col_heading { color: #333333; background-color: #f4f4f9; font-size: 100%; font-weight: normal; text-transform: inherit; border-left-style: none; border-left-width: 1px; border-left-color: #D3D3D3; border-right-style: none; border-right-width: 1px; border-right-color: #D3D3D3; vertical-align: bottom; padding-top: 5px; padding-bottom: 6px; padding-left: 5px; padding-right: 5px; overflow-x: hidden; } #kfibdwimrn .gt_column_spanner_outer { color: #333333; background-color: #f4f4f9; font-size: 100%; font-weight: normal; text-transform: inherit; padding-top: 0; padding-bottom: 0; padding-left: 4px; padding-right: 4px; } #kfibdwimrn .gt_column_spanner_outer:first-child { padding-left: 0; } #kfibdwimrn .gt_column_spanner_outer:last-child { padding-right: 0; } #kfibdwimrn .gt_column_spanner { border-bottom-style: solid; border-bottom-width: 2px; border-bottom-color: #D3D3D3; vertical-align: bottom; padding-top: 5px; padding-bottom: 6px; overflow-x: hidden; display: inline-block; width: 100%; } #kfibdwimrn .gt_group_heading { padding: 8px; color: #333333; background-color: #f4f4f9; font-size: 100%; font-weight: initial; text-transform: inherit; border-top-style: solid; border-top-width: 2px; border-top-color: #D3D3D3; border-bottom-style: solid; border-bottom-width: 2px; border-bottom-color: #D3D3D3; border-left-style: none; border-left-width: 1px; border-left-color: #D3D3D3; border-right-style: none; border-right-width: 1px; border-right-color: #D3D3D3; vertical-align: middle; } #kfibdwimrn .gt_empty_group_heading { padding: 0.5px; color: #333333; background-color: #f4f4f9; font-size: 100%; font-weight: initial; border-top-style: solid; border-top-width: 2px; border-top-color: #D3D3D3; border-bottom-style: solid; border-bottom-width: 2px; border-bottom-color: #D3D3D3; vertical-align: middle; } #kfibdwimrn .gt_from_md > :first-child { margin-top: 0; } #kfibdwimrn .gt_from_md > :last-child { margin-bottom: 0; } #kfibdwimrn .gt_row { padding-top: 8px; padding-bottom: 8px; padding-left: 5px; padding-right: 5px; margin: 10px; border-top-style: solid; border-top-width: 1px; border-top-color: #D3D3D3; border-left-style: none; border-left-width: 1px; border-left-color: #D3D3D3; border-right-style: none; border-right-width: 1px; border-right-color: #D3D3D3; vertical-align: middle; overflow-x: hidden; } #kfibdwimrn .gt_stub { color: #333333; background-color: #f4f4f9; font-size: 100%; font-weight: initial; text-transform: inherit; border-right-style: solid; border-right-width: 2px; border-right-color: #D3D3D3; padding-left: 12px; } #kfibdwimrn .gt_summary_row { color: #333333; background-color: #f4f4f9; text-transform: inherit; padding-top: 8px; padding-bottom: 8px; padding-left: 5px; padding-right: 5px; } #kfibdwimrn .gt_first_summary_row { padding-top: 8px; padding-bottom: 8px; padding-left: 5px; padding-right: 5px; border-top-style: solid; border-top-width: 2px; border-top-color: #D3D3D3; } #kfibdwimrn .gt_grand_summary_row { color: #333333; background-color: #f4f4f9; text-transform: inherit; padding-top: 8px; padding-bottom: 8px; padding-left: 5px; padding-right: 5px; } #kfibdwimrn .gt_first_grand_summary_row { padding-top: 8px; padding-bottom: 8px; padding-left: 5px; padding-right: 5px; border-top-style: double; border-top-width: 6px; border-top-color: #D3D3D3; } #kfibdwimrn .gt_striped { background-color: rgba(128, 128, 128, 0.05); } #kfibdwimrn .gt_table_body { border-top-style: solid; border-top-width: 2px; border-top-color: #D3D3D3; border-bottom-style: solid; border-bottom-width: 2px; border-bottom-color: #D3D3D3; } #kfibdwimrn .gt_footnotes { color: #333333; background-color: #f4f4f9; border-bottom-style: none; border-bottom-width: 2px; border-bottom-color: #D3D3D3; border-left-style: none; border-left-width: 2px; border-left-color: #D3D3D3; border-right-style: none; border-right-width: 2px; border-right-color: #D3D3D3; } #kfibdwimrn .gt_footnote { margin: 0px; font-size: 90%; padding: 4px; } #kfibdwimrn .gt_sourcenotes { color: #333333; background-color: #f4f4f9; border-bottom-style: none; border-bottom-width: 2px; border-bottom-color: #D3D3D3; border-left-style: none; border-left-width: 2px; border-left-color: #D3D3D3; border-right-style: none; border-right-width: 2px; border-right-color: #D3D3D3; } #kfibdwimrn .gt_sourcenote { font-size: 90%; padding: 4px; } #kfibdwimrn .gt_left { text-align: left; } #kfibdwimrn .gt_center { text-align: center; } #kfibdwimrn .gt_right { text-align: right; font-variant-numeric: tabular-nums; } #kfibdwimrn .gt_font_normal { font-weight: normal; } #kfibdwimrn .gt_font_bold { font-weight: bold; } #kfibdwimrn .gt_font_italic { font-style: italic; } #kfibdwimrn .gt_super { font-size: 65%; } #kfibdwimrn .gt_footnote_marks { font-style: italic; font-size: 65%; } </style> <div id="kfibdwimrn" style="overflow-x:auto;overflow-y:auto;width:auto;height:auto;"><table class="gt_table"> <thead class="gt_col_headings"> <tr> <th class="gt_col_heading gt_columns_bottom_border gt_left" rowspan="1" colspan="1" style="font-size: 21px;">Sample</th> <th class="gt_col_heading gt_columns_bottom_border gt_right" rowspan="1" colspan="1" style="font-size: 21px;">Main.Sensor</th> <th class="gt_col_heading gt_columns_bottom_border gt_right" rowspan="1" colspan="1" style="font-size: 21px;">Backup.Sensor</th> <th class="gt_col_heading gt_columns_bottom_border gt_right" rowspan="1" colspan="1" style="font-size: 21px;">Manual.Reading</th> </tr> </thead> <tbody class="gt_table_body"> <tr><td class="gt_row gt_left" style="font-size: 21px;">A</td> <td class="gt_row gt_right" style="font-size: 21px;">NA</td> <td class="gt_row gt_right" style="font-size: 21px;">12.1</td> <td class="gt_row gt_right" style="font-size: 21px;">12.0</td></tr> <tr><td class="gt_row gt_left" style="font-size: 21px;">B</td> <td class="gt_row gt_right" style="font-size: 21px;">12</td> <td class="gt_row gt_right" style="font-size: 21px;">NA</td> <td class="gt_row gt_right" style="font-size: 21px;">11.9</td></tr> <tr><td class="gt_row gt_left" style="font-size: 21px;">C</td> <td class="gt_row gt_right" style="font-size: 21px;">16</td> <td class="gt_row gt_right" style="font-size: 21px;">NA</td> <td class="gt_row gt_right" style="font-size: 21px;">NA</td></tr> <tr><td class="gt_row gt_left" style="font-size: 21px;">D</td> <td class="gt_row gt_right" style="font-size: 21px;">3</td> <td class="gt_row gt_right" style="font-size: 21px;">9.5</td> <td class="gt_row gt_right" style="font-size: 21px;">NA</td></tr> <tr><td class="gt_row gt_left" style="font-size: 21px;">E</td> <td class="gt_row gt_right" style="font-size: 21px;">NA</td> <td class="gt_row gt_right" style="font-size: 21px;">13.0</td> <td class="gt_row gt_right" style="font-size: 21px;">12.0</td></tr> </tbody> </table></div> ] .panel[.panel-name[Data setup] ```r sensors <- tibble::tribble( ~Sample, ~Main.Sensor, ~Backup.Sensor, ~Manual.Reading, "A", NA, 12.1, 12, "B", 12L, NA, 11.9, "C", 16L, NA, NA, "D", 3L, 9.5, NA, "E", NA, 13, 12 ) ``` ] ] ??? These data has temperature readings for different samples from two automated sensors plus a manual reading with an external device. For whatever season, there are missing values in the main sensor data, so we can coalesce the main, backup, and manual readings into a single column. These data have an inherent priority and order, in which we want to fill in gaps in the main sensor data with data from the backup sensor, with the manual readings as a third option. --- </br> ```r sensors %>% mutate(Final.Reading=coalesce(Main.Sensor, Backup.Sensor, Manual.Reading)) ``` ``` ## # A tibble: 5 x 5 ## Sample Main.Sensor Backup.Sensor Manual.Reading Final.Reading ## <chr> <int> <dbl> <dbl> <dbl> ## 1 A NA 12.1 12 12.1 ## 2 B 12 NA 11.9 12 ## 3 C 16 NA NA 16 ## 4 D 3 9.5 NA 3 ## 5 E NA 13 12 13 ``` ??? The code would look like this, we use mutate to create a new column called final reading, which is the output from coalescing the three columns with the data. In this case, we specified the same order that they appear in in the data. The new column combines data from the first column with values from the second one, and the third one was not needed. --- class: center  ??? This animation might also show the process better. If we have a record of the process and we don’t overwrite our original data, we may even want to subset the data to keep only the new column. --- class: center, middle, dk-section-title background-image:url("images/sen-sathyamony-4RkmVyBMO5Q-unsplash.jpg") background-size: cover # Filling Adjacent Values ??? To wrap up, Let’s see how we can fill adjacent values in R. Doing this helps us avoid unnecessary NA values and makes for less ambiguous data. The logic is similar to the one you may be familiar with from filling adjacent values in spreadsheet software. --- ## .b.orange[fill()] from 📦 .b.rrured[`tidyr`] ??? We’ll use the fill function from the tidyr package. The usage is straight forward. -- .fl.w-40[ <style>html { font-family: -apple-system, BlinkMacSystemFont, 'Segoe UI', Roboto, Oxygen, Ubuntu, Cantarell, 'Helvetica Neue', 'Fira Sans', 'Droid Sans', Arial, sans-serif; } #itjyfntzls .gt_table { display: table; border-collapse: collapse; margin-left: auto; margin-right: auto; color: #333333; font-size: 16px; font-weight: normal; font-style: normal; background-color: #f4f4f9; width: auto; border-top-style: solid; border-top-width: 2px; border-top-color: #A8A8A8; border-right-style: none; border-right-width: 2px; border-right-color: #D3D3D3; border-bottom-style: solid; border-bottom-width: 2px; border-bottom-color: #A8A8A8; border-left-style: none; border-left-width: 2px; border-left-color: #D3D3D3; } #itjyfntzls .gt_heading { background-color: #f4f4f9; text-align: center; border-bottom-color: #f4f4f9; border-left-style: none; border-left-width: 1px; border-left-color: #D3D3D3; border-right-style: none; border-right-width: 1px; border-right-color: #D3D3D3; } #itjyfntzls .gt_title { color: #333333; font-size: 125%; font-weight: initial; padding-top: 4px; padding-bottom: 4px; border-bottom-color: #f4f4f9; border-bottom-width: 0; } #itjyfntzls .gt_subtitle { color: #333333; font-size: 85%; font-weight: initial; padding-top: 0; padding-bottom: 4px; border-top-color: #f4f4f9; border-top-width: 0; } #itjyfntzls .gt_bottom_border { border-bottom-style: solid; border-bottom-width: 2px; border-bottom-color: #D3D3D3; } #itjyfntzls .gt_col_headings { border-top-style: solid; border-top-width: 2px; border-top-color: #D3D3D3; border-bottom-style: solid; border-bottom-width: 2px; border-bottom-color: #D3D3D3; border-left-style: none; border-left-width: 1px; border-left-color: #D3D3D3; border-right-style: none; border-right-width: 1px; border-right-color: #D3D3D3; } #itjyfntzls .gt_col_heading { color: #333333; background-color: #f4f4f9; font-size: 100%; font-weight: normal; text-transform: inherit; border-left-style: none; border-left-width: 1px; border-left-color: #D3D3D3; border-right-style: none; border-right-width: 1px; border-right-color: #D3D3D3; vertical-align: bottom; padding-top: 5px; padding-bottom: 6px; padding-left: 5px; padding-right: 5px; overflow-x: hidden; } #itjyfntzls .gt_column_spanner_outer { color: #333333; background-color: #f4f4f9; font-size: 100%; font-weight: normal; text-transform: inherit; padding-top: 0; padding-bottom: 0; padding-left: 4px; padding-right: 4px; } #itjyfntzls .gt_column_spanner_outer:first-child { padding-left: 0; } #itjyfntzls .gt_column_spanner_outer:last-child { padding-right: 0; } #itjyfntzls .gt_column_spanner { border-bottom-style: solid; border-bottom-width: 2px; border-bottom-color: #D3D3D3; vertical-align: bottom; padding-top: 5px; padding-bottom: 6px; overflow-x: hidden; display: inline-block; width: 100%; } #itjyfntzls .gt_group_heading { padding: 8px; color: #333333; background-color: #f4f4f9; font-size: 100%; font-weight: initial; text-transform: inherit; border-top-style: solid; border-top-width: 2px; border-top-color: #D3D3D3; border-bottom-style: solid; border-bottom-width: 2px; border-bottom-color: #D3D3D3; border-left-style: none; border-left-width: 1px; border-left-color: #D3D3D3; border-right-style: none; border-right-width: 1px; border-right-color: #D3D3D3; vertical-align: middle; } #itjyfntzls .gt_empty_group_heading { padding: 0.5px; color: #333333; background-color: #f4f4f9; font-size: 100%; font-weight: initial; border-top-style: solid; border-top-width: 2px; border-top-color: #D3D3D3; border-bottom-style: solid; border-bottom-width: 2px; border-bottom-color: #D3D3D3; vertical-align: middle; } #itjyfntzls .gt_from_md > :first-child { margin-top: 0; } #itjyfntzls .gt_from_md > :last-child { margin-bottom: 0; } #itjyfntzls .gt_row { padding-top: 8px; padding-bottom: 8px; padding-left: 5px; padding-right: 5px; margin: 10px; border-top-style: solid; border-top-width: 1px; border-top-color: #D3D3D3; border-left-style: none; border-left-width: 1px; border-left-color: #D3D3D3; border-right-style: none; border-right-width: 1px; border-right-color: #D3D3D3; vertical-align: middle; overflow-x: hidden; } #itjyfntzls .gt_stub { color: #333333; background-color: #f4f4f9; font-size: 100%; font-weight: initial; text-transform: inherit; border-right-style: solid; border-right-width: 2px; border-right-color: #D3D3D3; padding-left: 12px; } #itjyfntzls .gt_summary_row { color: #333333; background-color: #f4f4f9; text-transform: inherit; padding-top: 8px; padding-bottom: 8px; padding-left: 5px; padding-right: 5px; } #itjyfntzls .gt_first_summary_row { padding-top: 8px; padding-bottom: 8px; padding-left: 5px; padding-right: 5px; border-top-style: solid; border-top-width: 2px; border-top-color: #D3D3D3; } #itjyfntzls .gt_grand_summary_row { color: #333333; background-color: #f4f4f9; text-transform: inherit; padding-top: 8px; padding-bottom: 8px; padding-left: 5px; padding-right: 5px; } #itjyfntzls .gt_first_grand_summary_row { padding-top: 8px; padding-bottom: 8px; padding-left: 5px; padding-right: 5px; border-top-style: double; border-top-width: 6px; border-top-color: #D3D3D3; } #itjyfntzls .gt_striped { background-color: rgba(128, 128, 128, 0.05); } #itjyfntzls .gt_table_body { border-top-style: solid; border-top-width: 2px; border-top-color: #D3D3D3; border-bottom-style: solid; border-bottom-width: 2px; border-bottom-color: #D3D3D3; } #itjyfntzls .gt_footnotes { color: #333333; background-color: #f4f4f9; border-bottom-style: none; border-bottom-width: 2px; border-bottom-color: #D3D3D3; border-left-style: none; border-left-width: 2px; border-left-color: #D3D3D3; border-right-style: none; border-right-width: 2px; border-right-color: #D3D3D3; } #itjyfntzls .gt_footnote { margin: 0px; font-size: 90%; padding: 4px; } #itjyfntzls .gt_sourcenotes { color: #333333; background-color: #f4f4f9; border-bottom-style: none; border-bottom-width: 2px; border-bottom-color: #D3D3D3; border-left-style: none; border-left-width: 2px; border-left-color: #D3D3D3; border-right-style: none; border-right-width: 2px; border-right-color: #D3D3D3; } #itjyfntzls .gt_sourcenote { font-size: 90%; padding: 4px; } #itjyfntzls .gt_left { text-align: left; } #itjyfntzls .gt_center { text-align: center; } #itjyfntzls .gt_right { text-align: right; font-variant-numeric: tabular-nums; } #itjyfntzls .gt_font_normal { font-weight: normal; } #itjyfntzls .gt_font_bold { font-weight: bold; } #itjyfntzls .gt_font_italic { font-style: italic; } #itjyfntzls .gt_super { font-size: 65%; } #itjyfntzls .gt_footnote_marks { font-style: italic; font-size: 65%; } </style> <div id="itjyfntzls" style="overflow-x:auto;overflow-y:auto;width:auto;height:auto;"><table class="gt_table"> <thead class="gt_col_headings"> <tr> <th class="gt_col_heading gt_columns_bottom_border gt_left" rowspan="1" colspan="1" style="font-size: 21px;">animal</th> <th class="gt_col_heading gt_columns_bottom_border gt_right" rowspan="1" colspan="1" style="font-size: 21px;">population</th> </tr> </thead> <tbody class="gt_table_body"> <tr><td class="gt_row gt_left" style="font-size: 21px;">Bird</td> <td class="gt_row gt_right" style="font-size: 21px;">21</td></tr> <tr><td class="gt_row gt_left" style="font-size: 21px;">NA</td> <td class="gt_row gt_right" style="font-size: 21px;">5</td></tr> <tr><td class="gt_row gt_left" style="font-size: 21px;">NA</td> <td class="gt_row gt_right" style="font-size: 21px;">7</td></tr> <tr><td class="gt_row gt_left" style="font-size: 21px;">NA</td> <td class="gt_row gt_right" style="font-size: 21px;">23</td></tr> <tr><td class="gt_row gt_left" style="font-size: 21px;">NA</td> <td class="gt_row gt_right" style="font-size: 21px;">74</td></tr> <tr><td class="gt_row gt_left" style="font-size: 21px;">Frog</td> <td class="gt_row gt_right" style="font-size: 21px;">23</td></tr> <tr><td class="gt_row gt_left" style="font-size: 21px;">NA</td> <td class="gt_row gt_right" style="font-size: 21px;">65</td></tr> <tr><td class="gt_row gt_left" style="font-size: 21px;">NA</td> <td class="gt_row gt_right" style="font-size: 21px;">12</td></tr> <tr><td class="gt_row gt_left" style="font-size: 21px;">NA</td> <td class="gt_row gt_right" style="font-size: 21px;">5</td></tr> </tbody> </table></div> ] </br> `animal_populations` ??? Look at this two column table with data on animal populations. To avoid repetition, values in the “animal” column only appear when they change to a different one. -- .large[- Fill missing values in a column (top to bottom by default)] .large[- Contiguous/adjacent values assumed to be the same] .large[- Values only appear when they change along a column] ??? Fill will copy the value down into missing values, assuming that contiguous values are the same, and will stop at the next non-missing value. --- ## fill() .pull-left[ ```r animal_populations ``` ``` ## # A tibble: 9 x 2 ## animal population ## <chr> <dbl> ## 1 Bird 21 ## 2 <NA> 5 ## 3 <NA> 7 ## 4 <NA> 23 ## 5 <NA> 74 ## 6 Frog 23 ## 7 <NA> 65 ## 8 <NA> 12 ## 9 <NA> 5 ``` ] .pull-right[ ```r animal_populations %>% fill(animal) ``` ``` ## # A tibble: 9 x 2 ## animal population ## <chr> <dbl> ## 1 Bird 21 ## 2 Bird 5 ## 3 Bird 7 ## 4 Bird 23 ## 5 Bird 74 ## 6 Frog 23 ## 7 Frog 65 ## 8 Frog 12 ## 9 Frog 5 ``` ] ??? We can specify which variable we want to fill, by name, as an argument to the fill function. Here we are filling the animal column, and we can see that the values were copied down, replacing the NAS. --- class: my-turn # My Turn -- .large[- Load the fish landings data 'fish-landings.csv'] -- .large[- Fill the 'Fish' and 'Lake' columns] -- .large[- Create a new column, coalescing the three numeric variables] --- class: inverse # Your Turn -- .large[- Load the fish landings data 'fish-landings.csv'] -- .large[- Fill the 'Fish' and 'Lake' columns] -- .large[- Reorder the numeric variables ('Commission reported total' first) and create a new column, coalescing the three numeric variables]